
| 标记 Marker | 染色体 Chromosome | 位置 Position/cM | 等位基因位点 Allelic loci | 基因型数 Genotype Number | 多态性信息含量 Polymorphic information content |
|---|---|---|---|---|---|
| HAU3193 | Chr01 | 21.224 | 1 | 2 | 0.088 |
| NAU3736b | Chr01 | 69.402 | 2 | 4 | 0.554 |
| MUSS422a | Chr01 | 95.112 | 2 | 5 | 0.378 |
| NBRI_HQ526730 | Chr01 | 133.007 | 3 | 8 | 0.660 |
| NBRI_HQ527566 | Chr01 | 150.217 | 1 | 2 | 0.373 |
| MON_CGR5007 | Chr02 | 0 | 2 | 3 | 0.090 |
| MON_SHIN-1481 | Chr02 | 78.766 | 2 | 3 | 0.196 |
| MON_SHIN-1584 | Chr02 | 101.604 | 2 | 3 | 0.411 |
| NAU3419c | Chr02 | 156.034 | 1 | 3 | 0.459 |
| HAU0875 | Chr03 | 31.868 | 1 | 3 | 0.090 |
| HAU2588 | Chr03 | 50.948 | 2 | 4 | 0.440 |
| NBRI_HQ527820 | Chr03 | 112.996 | 4 | 9 | 0.582 |
| CK988221-PR-ss | Chr03 | 147.159 | 1 | 3 | 0.344 |
| NAU7182 | Chr04 | 33.344 | 1 | 3 | 0.366 |
| HAU3371 | Chr04 | 72.457 | 1 | 3 | 0.306 |
| Gh107 | Chr04 | 105.848 | 1 | 3 | 0.370 |
| BNL2865 | Chr05 | 25.983 | 1 | 3 | 0.422 |
| MON_CGR5732 | Chr05 | 72.914 | 2 | 5 | 0.562 |
| HAU1952b | Chr05 | 116.03 | 4 | 10 | 0.540 |
| MON_DPL0024 | Chr05 | 211.259 | 2 | 5 | 0.350 |
| HAU1496 | Chr05 | 231.991 | 1 | 3 | 0.104 |
| MON_DPL0375c | Chr06 | 43.592 | 3 | 9 | 0.732 |
| DPL0238 | Chr06 | 52.86 | 2 | 5 | 0.546 |
| MON_DPL0811b | Chr06 | 105.485 | 2 | 5 | 0.480 |
| MON_SHIN-1585 | Chr07 | 13.29 | 2 | 5 | 0.121 |
| HAU-SNP113 | Chr07 | 72.462 | 2 | 5 | 0.571 |
| NAU6734 | Chr08 | 4.383 | 1 | 3 | 0.145 |
| MON_SHIN-1494b | Chr08 | 108.27 | 1 | 3 | 0.209 |
| BNL3031 | Chr09 | 108.086 | 2 | 5 | 0.650 |
| NAU5323 | Chr10 | 0 | 1 | 2 | 0.088 |
| HAU-SNP085 | Chr10 | 143.325 | 2 | 4 | 0.249 |
| MON_SHIN-0337a | Chr11 | 162.89 | 1 | 3 | 0.508 |
| MGHES31 | Chr12 | 52.488 | 3 | 7 | 0.564 |
| MON_DPL0010 | Chr12 | 109.666 | 2 | 4 | 0.527 |
| MON_CGR6012 | Chr12 | 114.01 | 2 | 5 | 0.675 |
| MON_CGR6812 | Chr13 | 5.883 | 2 | 5 | 0.694 |
| BNL2449 | Chr13 | 80.886 | 2 | 4 | 0.646 |
| HAU4748 | Chr13 | 204.131 | 2 | 5 | 0.550 |
| MON_DPL0521 | Chr14 | 17.211 | 2 | 4 | 0.134 |
| MON_DPL0502 | Chr14 | 32.554 | 3 | 7 | 0.679 |
| NAU4022 | Chr14 | 48.815 | 1 | 2 | 0.046 |
| NAU3913 | Chr14 | 71.987 | 3 | 7 | 0.453 |
| HAU3071a | Chr14 | 159.457 | 4 | 12 | 0.699 |
| NAU3346b | Chr15 | 0 | 3 | 7 | 0.606 |
| NAU5172 | Chr15 | 89.91 | 2 | 4 | 0.149 |
| NAU2985 | Chr15 | 119.125 | 2 | 5 | 0.473 |
| NAU3433 | Chr15 | 125.561 | 2 | 5 | 0.432 |
| NAU7049 | Chr15 | 161.039 | 2 | 5 | 0.451 |
| HAU2481 | Chr16 | 69.382 | 2 | 5 | 0.343 |
| HAU4022 | Chr17 | 102.33 | 3 | 5 | 0.295 |
| NAU3827 | Chr18 | 120.959 | 2 | 3 | 0.077 |
| MON_DPL0893 | Chr19 | 22.741 | 2 | 4 | 0.175 |
| HAU4483 | Chr19 | 76.798 | 2 | 3 | 0.309 |
| NAU2126 | Chr19 | 147.806 | 2 | 5 | 0.312 |
| HAU2846 | Chr19 | 227.882 | 3 | 7 | 0.365 |
| BNL3043 | Chr19 | 238.658 | 2 | 5 | 0.493 |
| HAU1968a | Chr20 | 25.402 | 1 | 2 | 0.074 |
| NAU6997 | Chr20 | 42.205 | 2 | 5 | 0.213 |
| MON_DPL0504a | Chr20 | 112.354 | 3 | 5 | 0.441 |
| CCRI596a | Chr21 | 18.404 | 3 | 6 | 0.165 |
| DPL0062 | Chr21 | 60.776 | 1 | 3 | 0.467 |
| HAU2770 | Chr21 | 107.624 | 2 | 5 | 0.372 |
| HAU0938 | Chr22 | 78.573 | 2 | 2 | 0.060 |
| Gh330 | Chr22 | 106.752 | 3 | 6 | 0.190 |
| BNL2535b | Chr23 | 40.99 | 2 | 4 | 0.197 |
| CCRI303 | Chr24 | 6.332 | 2 | 5 | 0.372 |
| MON_CGR5447 | Chr24 | 74.443 | 2 | 5 | 0.564 |
| NAU2240 | Chr24 | 118.855 | 2 | 4 | 0.147 |
| NAU2631 | Chr24 | 140.64 | 2 | 5 | 0.310 |
| NAU2713a | Chr25 | 7.701 | 2 | 4 | 0.338 |
| NAU3298 | Chr25 | 48.872 | 2 | 5 | 0.107 |
| NAU3774 | Chr26 | 55.717 | 2 | 5 | 0.510 |
| NAU4089 | Chr26 | 180.828 | 1 | 3 | 0.118 |
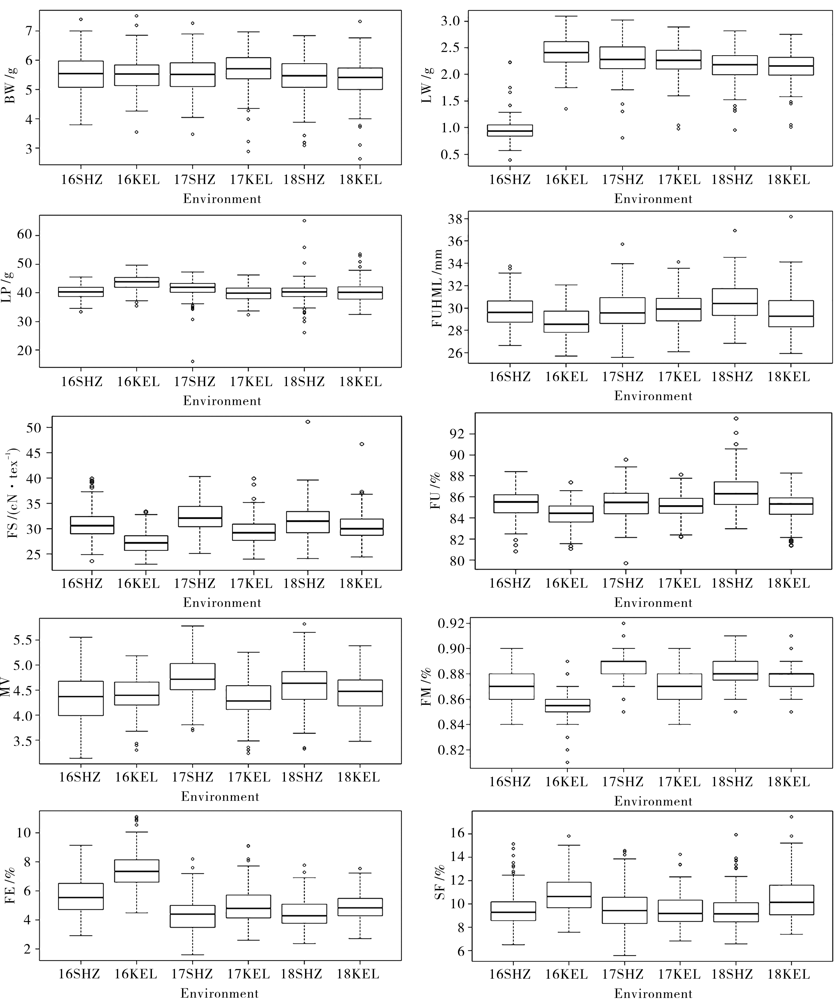 Fig. 1 Boxplot diagram of 10 traits in Shihezi and Korla regions from 2016 to 2018 The ends of boxplot, the circles out of both ends of boxplot, and the bold lines in the middle show the extreme range, single the maximum and minimum value, and the mean of phenotypic value of the targeted trait, respectively.
Fig. 1 Boxplot diagram of 10 traits in Shihezi and Korla regions from 2016 to 2018 The ends of boxplot, the circles out of both ends of boxplot, and the bold lines in the middle show the extreme range, single the maximum and minimum value, and the mean of phenotypic value of the targeted trait, respectively.